Method that identify species from bulk sample DNA or environmental DNA via high-throughput sequencing of a DNA marker is known as DNA metabarcoding. Any gene or DNA fragment that is used to identify a species, individual or genotype is known as DNA marker.
DNA metabarcoding is a rapid method of biodiversity assessment that combines two technologies: DNA based identification and high-throughput DNA sequencing. The simultaneous sequencing of millions of DNA fragments is known as High-throughput sequencing (HTS).
How DNA metabarcoding is different from DNA barcoding?
Also refer DNA barcoding https://fotisedu.com/dna-barcoding/
DNA metabarcoding infers the species composition of an environmental sample by amplifying, sequencing, and analysing target genomic regions. It differs from DNA barcoding in the use of high-throughput sequencing. This technique allows for DNA sequencing of bulk samples without a prior step of specimen sorting. DNA barcode is a DNA marker routinely used for identifying species – e.g. the mitochondrial cytochrome c oxidase I (COI) gene is the standard DNA barcode for identifying animal species.
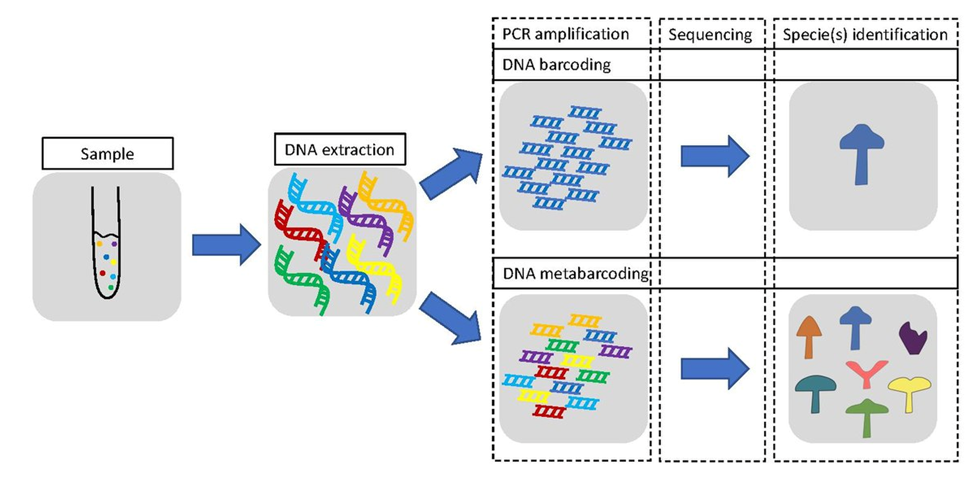
With DNA metabarcoding we can identify organisms down to various taxonomic levels and compare the taxa composition among samples. DNA metabarcoding can be used even when DNA is degraded. Therefore, it is possible to analyse taxa diversity in samples such as soil, faeces, or sediments.
Reasons for increasing popularity of DNA metabarcoding
DNA metabarcoding is rapidly emerging as a cost-effective approach for large-scale studies sampling environments where constraints of conventional morphology-based species identification is logistically or financially impractical. Invertebrates are among the most studied taxa in DNA metabarcoding studies.
Environmental DNA (eDNA) is the DNA extracted from environmental samples, such as soil, water, or sediment, without prior isolation of target organisms
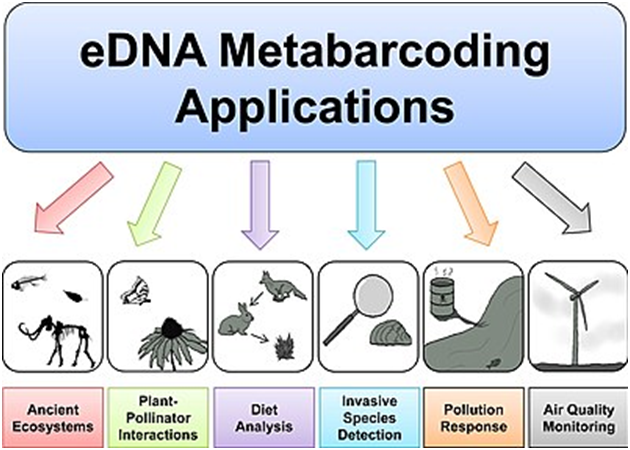
There is an increasing demand for this technology in a variety of fields, such as:
1 . Microbial ecology, for characterising microbial communities from several types of samples (water, soil, air,etc.).
2 . Aerobiology, for identifying organisms present in air samples, like bacteria, fungal spores, and pollen.
3 . Feeding ecology, where prey species can be identified by analysing DNA from the predators’ faeces.
4 . Soil biology, for identifying organisms (bacteria, fungi, small animals,etc.) present in soil samples.
5 . Ecosystem monitoring, for screening bioindicator species present in several types of samples.
6 . Marine and freshwater biology, for identifying microalgae and larvae present in water samples.
